Normalizing a cancer contact count matrix with CAIC¶
CAIC is a normalization method to remove copy-number biases present in a matrix. This example showcases how to perform such a normalization on simulated data with iced.
Loading the data and normalizing¶
The normalization is done in three step:
- Normalize the data using LOIC, to remove GC, mappability, and other biases
- Estimate the block biases due to copy number.
- Remove the block biases from the LOIC-normalized contact counts
from iced import datasets
from iced import normalization
import matplotlib.pyplot as plt
from matplotlib import colors
counts, lengths, cnv = datasets.load_sample_cancer()
loic_normed = normalization.ICE_normalization(counts, counts_profile=cnv)
block_biases = normalization.estimate_block_biases(counts, lengths, cnv)
caic_normed = loic_normed / block_biases
Out:
Estimating CNV-effects.
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
Computing relationship genomic distance & expected counts
Fitting Isotonic Regression
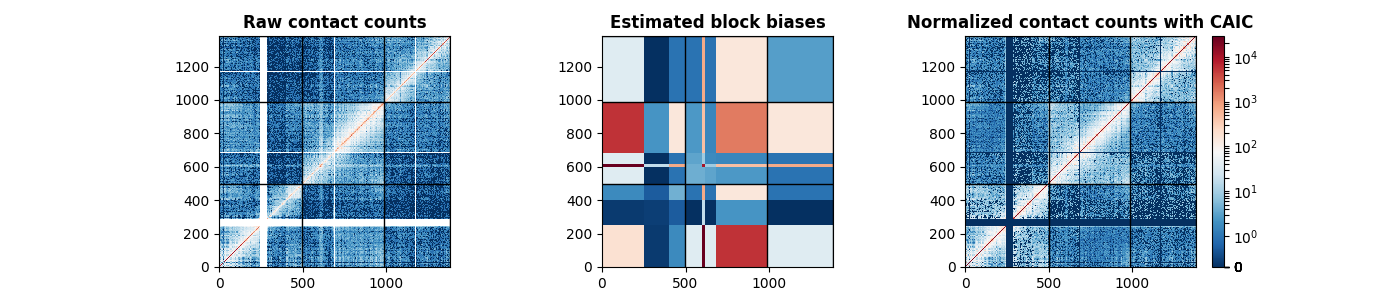
Visualizing the results using Matplotlib¶
The following code visualizes the raw original data, the estimated block biases, and the normalized matrix using the CAIC method.
chromosomes = ["I", "II", "III", "IV", "V", "VI"]
fig, axes = plt.subplots(ncols=3, figsize=(14, 3))
axes[0].imshow(counts, cmap="RdBu_r", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
[axes[0].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[0].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
axes[0].set_title("Raw contact counts", fontweight="bold")
m = axes[1].imshow(block_biases, cmap="RdBu_r", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
[axes[1].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[1].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
axes[1].set_title("Estimated block biases", fontweight="bold")
m = axes[2].imshow(caic_normed,
cmap="RdBu_r", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
[axes[2].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[2].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
cb = fig.colorbar(m)
axes[2].set_title("Normalized contact counts with CAIC", fontweight="bold")
Total running time of the script: ( 0 minutes 24.015 seconds)