Normalizing a contact count matrix¶
This example showcases some basic filtering and normalization.
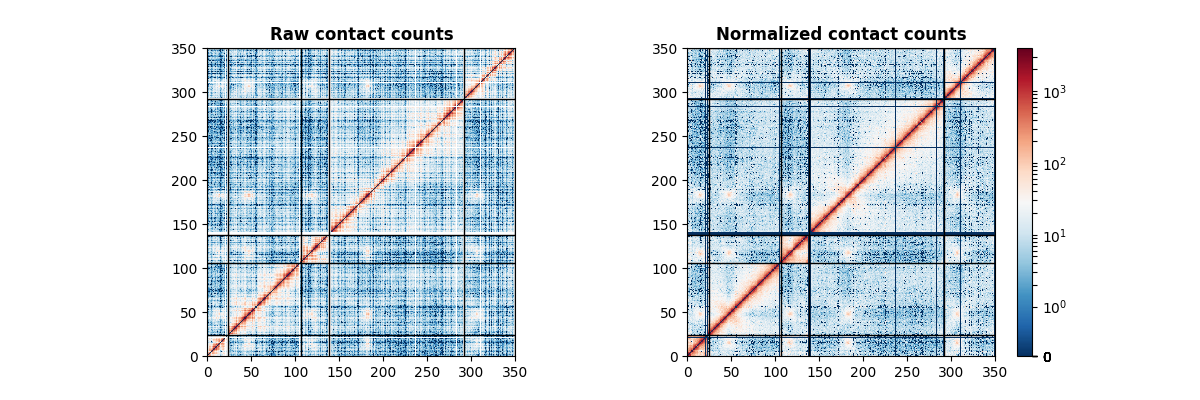
import matplotlib.pyplot as plt
from matplotlib import colors
from iced import datasets
from iced import filter
from iced import normalization
# Loading a sample dataset
counts, lengths = datasets.load_sample_yeast()
# Filtering and normalizing contact count data
normed = filter.filter_low_counts(counts, lengths=lengths, percentage=0.04)
normed = normalization.ICE_normalization(normed)
# Plotting the results using matplotlib
chromosomes = ["I", "II", "III", "IV", "V", "VI"]
fig, axes = plt.subplots(ncols=2, figsize=(12, 4))
axes[0].imshow(counts, cmap="RdBu_r", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
[axes[0].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[0].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
axes[0].set_title("Raw contact counts", fontweight="bold")
m = axes[1].imshow(normed, cmap="RdBu_r", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
[axes[1].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[1].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
cb = fig.colorbar(m)
axes[1].set_title("Normalized contact counts", fontweight="bold")
Total running time of the script: ( 0 minutes 0.403 seconds)