Extracting parts of a contact map¶
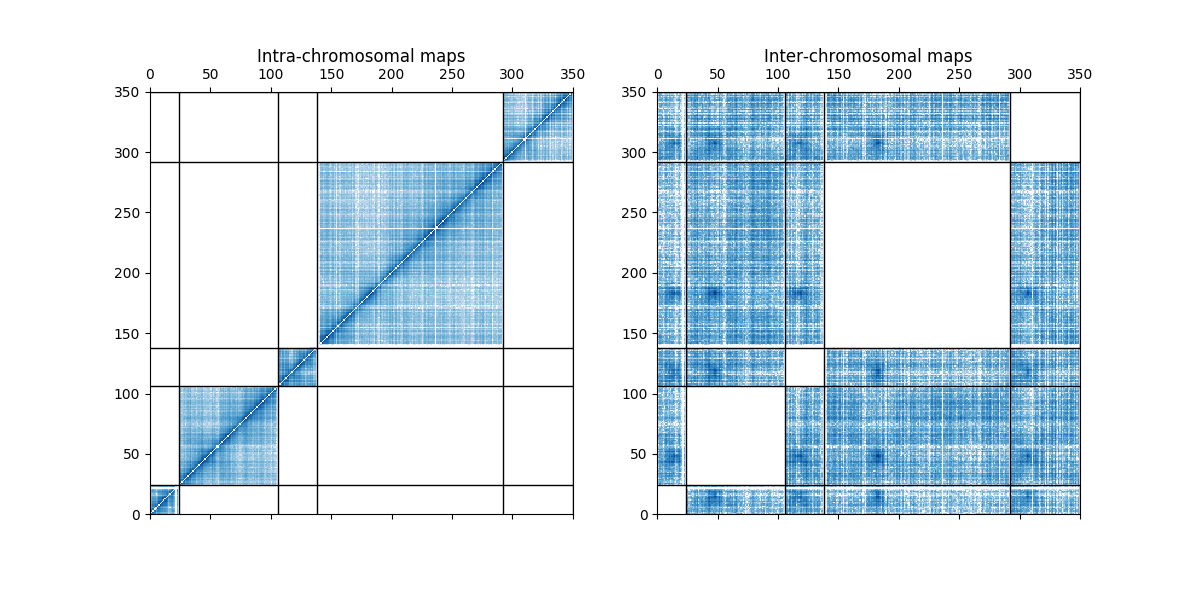
This examples shows how to use a mask to plot only the inter or the intra contact map.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import colors
from iced import datasets
from iced.utils import get_intra_mask
from iced.utils import get_inter_mask
# Loading a sample dataset
counts, lengths = datasets.load_sample_yeast()
intra_mask = get_intra_mask(lengths)
inter_mask = get_inter_mask(lengths)
fig, axes = plt.subplots(ncols=2, figsize=(12, 6))
inter_counts = counts.copy()
inter_counts[intra_mask] = np.nan
intra_counts = counts.copy()
intra_counts[inter_mask] = np.nan
m = axes[0].matshow(intra_counts, cmap="Blues", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
m = axes[1].matshow(inter_counts, cmap="Blues", norm=colors.SymLogNorm(1),
origin="bottom",
extent=(0, len(counts), 0, len(counts)))
axes[0].set_title("Intra-chromosomal maps")
axes[1].set_title("Inter-chromosomal maps")
[axes[0].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[0].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[1].axhline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
[axes[1].axvline(i, linewidth=1, color="#000000") for i in lengths.cumsum()]
Total running time of the script: ( 0 minutes 0.161 seconds)